Local software installation instructions for Bayes Workshop 2024
I recommend using Posit Cloud for this workshop, a demo server with all needed packages, software, and data installed (signup instructions here), especially if you have never used R before. If you decide not to use Posit Cloud for the workshop, but instead you want to install the software on your own machine, please follow the steps below. Note that after the workshop, if you want to analyze your own data, you’ll have to do this anyway or use Scinet. The Posit Cloud server is only intended for demonstration and teaching purposes. It is not a secure server, and government-owned data may not be uploaded to it.
NOTE: Some USDA employees have experienced issues getting this “software stack” working on their USDA Windows machines. Please contact me if you experience any problems, and copy and paste whatever error messages you get into the email.
My preferred setup for Bayesian models in R is to use the brms modeling package. brms is an R interface that allows you to write multilevel models with simple code that has similar syntax to the common mixed-model package lme4. “Behind the scenes” it fits a Bayesian model with the software Stan that has a state-of-the-art algorithm coded in C++ for quickly and efficiently sampling posterior distributions. There are two different packages that integrate between Stan and R, and you can specify which one to use in brms. While the default is rstan, the best option which makes the models run the fastest is CmdStanR. So in order to get brms working in R, you need to not only install brms but also have Stan installed on your system, and optionally CmdStanR. Then you have to set up everything so that all those pieces of software can communicate with each other.
I want to give props to Paul Bürkner, the developer of brms, and all the developers of Stan including Andrew Gelman, Bob Carpenter, and lots of other people. Their hard work makes our life easier and our stats better!
You’ll need to install the following software on your local machine.
- R (version 4.4)
- RStudio (optional but highly recommended)
- R packages
- brms
- cmdstanr
- agridat has the example datasets we’ll be using
- tidyverse for data manipulation and plotting
- emmeans for estimating means and testing hypotheses
- bayestestR for Bayesian hypothesis testing functions (part of the easystats package collection)
- tidybayes for extracting the results of Bayesian models into data objects that are easier to work with
- ggdist for creating beautiful plots of Bayesian model results
- gt for creating beautiful tables of Bayesian model results
- ape for calculating spatial autocorrelation statistics
- devtools needed behind the scenes
- Stan
Follow these steps to install R, RStudio, the needed R packages, and Stan.
NOTE: You will need to be connected to the Internet to install this software. I recommend disconnecting from any VPN before following these steps.
Step 1. Install R
You will need to download R from The Comprehensive R Archive Network, or CRAN. If you are using an older version of R, please update to version 4.4 or higher.
Go to the CRAN website and select the link to download the most recent version of R for your operating system. Follow the installation instructions. You will need admin rights to do this so you may need to get help from your IT staff.
Step 2. Install RStudio
RStudio is an additional software program that makes it easy for you to write R code. It provides an interface that lets you run code, edit scripts, see what variables are in your working environment, and manage files. RStudio is a program that makes it easier to use R, but you need to download both R and RStudio separately.

RStudio is now distributed by a company called Posit. Go to the RStudio Desktop download page and download the free version of RStudio Desktop. Again, you will need admin rights to do this on your USDA machine.
Step 3. Install R packages
Run the following code in your R or RStudio console (copy and paste). This installs all R packages needed for the workshop. Most are installed from the R package repository, CRAN. The cmdstanr package is installed from its own repository.
install.packages(c('brms', 'devtools', 'agridat', 'tidyverse', 'emmeans', 'tidybayes', 'ggdist', 'bayestestR', 'gt', 'ape'))
install.packages("cmdstanr", repos = c('https://stan-dev.r-universe.dev', getOption("repos")))
Step 4. Install CmdStan
If you don’t want to bother with CmdStan, skip down to the Testing the installation section below to test whether brms and Stan were installed correctly. But if you want to install CmdStan on your local machine, follow these CmdStan installation instructions.
Step 5. Test the installation
Run the following code in your R console. If you decided to skip installing CmdStan, omit the options(brms.backend = 'cmdstanr') line.
library(brms)
options(brms.backend = 'cmdstanr')
brm(mpg ~ hp, data = mtcars)
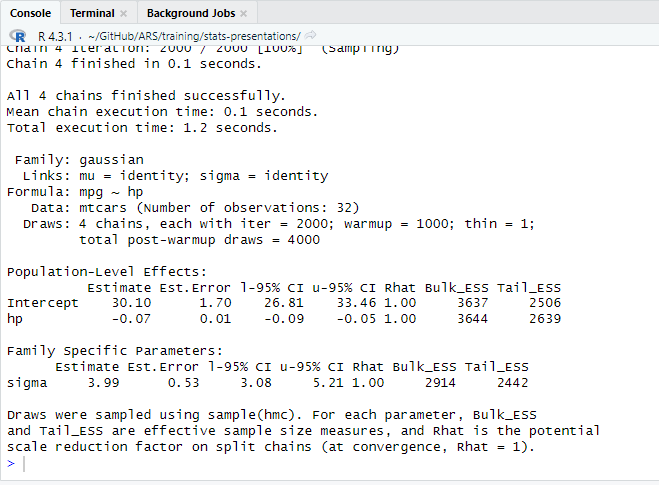
The above code loads brms and tells it to fit a simple model on a built-in example dataset. If it gives you a message that it is Compiling Stan model..., then progress indicators for the sampler, and then model summary output, as in the screenshot shown here, it means everything is working!
Step 6. Download the worksheet
Go back to the workshop homepage and download the worksheet. Open it in R or RStudio to follow along with the code during the workshop. Good luck!